Simulate sample heterogeneity
Sample heterogeneity is characterized by the presence of molecules with different characteristics in the data set. It can be simulated for various characteristics by using one of the following methods:
Parameter deviations
This method produces a continuous sample heterogeneity, i.e., molecule characteristics are broadened. This is opposed to the creation of sub-populations of molecules with distinguishable characteristics.
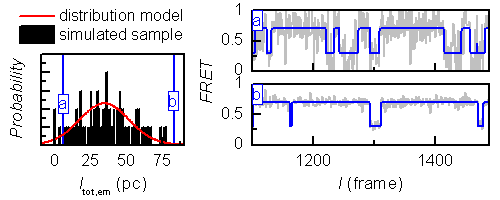
Sample heterogeneity can be introduced in FRET states FRETj, donor emission in absence of acceptor Itot,em and gamma factors γ.
Parameters are randomly generated for each molecule by drawing from Gaussian distributions with means respectively defined in fields
FRETj, Itot,em, γ, and standard deviation in fields wFRETj, wItot,em and wγ of the graphical interface.
Itot,em can be used to introduce heterogeneity in signal-to-noise ratio as shown in the following figure.
Molecule-specific pre-sets
This method allows to custom the type of sample heterogeneity by either (1) producing a broadening of molecule characteristics, or (2) creating sub-populations of molecules with distinguishable characteristics, or (3) both.
Sample heterogeneity can be introduced in FRET states FRETj, state transition rates kjj’, donor emission in absence of acceptor Itot,em, gamma factors γ and PSF widths wdet.
Parameters are pre-defined for individual molecules in a pre-set parameter file containing the respective variables FRET, trans_rates, tot_intensity, gamma and/or psf_width.
For more information about the structure and use of pre-set parameter file, please refer to
Pre-set parameters.
As an example, the script below creates two distinct molecule sub-populations of equal size, with one population allowing step-wise and reversible state transitions, and one allowing reversible transitions from state 1 only:
%%%%% EDIT HERE %%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
% defines 4-by-4 transition rates for the first population
trans_rates1 = [ 0 0.1 0 0
0.1 0 0.1 0
0 0.1 0 0.1
0 0 0.1 0];
% defines 4-by-4 transition rates for the second population
trans_rates2 = [ 0 0.1 0.1 0.1
0.1 0 0 0
0.1 0 0 0
0.1 0 0 0];
% defines the number of trajectories to simulate
N = 60;
% replicate the first and second transition rates matrix for half of the molecules with the repmat function
trans_rates1 = repmat(trans_rates1,[1 1 N/2]);
trans_rates2 = repmat(trans_rates2,[1 1 N/2]);
% concatenate both sub-populations into one sample
trans_rates = cat(3,trans_rates1,trans_rates2);
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
To build and export the pre-set parameter file:
-
Open the pre-set template in MATLAB’s editor by going to MATLAB’s menu
HOME → Open → Open ...and selecting the fileMASH-FRET/tools/createSimPrm.m. -
Define the desired variables in the corresponding section of the template (
trans_ratesfor the sub-mentioned example) -
Run the script by pressing
Runin MATLAB’s menuEDITORand export the pre-set file. -
load the pre-sets by pressing
 in
Pre-set parameters and selecting the corresponding file
in
Pre-set parameters and selecting the corresponding file