Transfer partial charges to mol2#
Here we will add partial charges from the AMBER force field (GROMACS residue topology files, rtp) to the RNA and DNA mononucleotides (mol2 files).
Processed nucleotide partial chages in FRETlabel on Github
input files:
DNA, RNA and aminoacid parameters from GROMACS rtp are located in
forcefields/1_amber_rtp/unprocessed (deoxy)nucleosides monophosphates in mol2 format are located in
fragments/1_bases/in/
output files:
processed (deoxy)nucleosides monophosphates mol2 files with updated partial charges will be located in
fragments/1_bases/out/
import pandas as pd
from biopandas.mol2 import PandasMol2
import fretlabel as fl
Load the deoxyribonucleotides mol2 files (with hydrogen names adapted to the AMBER ff) along with the corresponding rtp entries from the AMBER force field.
DA_mol2 = PandasMol2().read_mol2('fragments/1_bases/in/deoxyadenosine.mol2')
DT_mol2 = PandasMol2().read_mol2('fragments/1_bases/in/deoxythymidine.mol2')
DG_mol2 = PandasMol2().read_mol2('fragments/1_bases/in/deoxyguanosine.mol2')
DC_mol2 = PandasMol2().read_mol2('fragments/1_bases/in/deoxycytidine.mol2')
RA_mol2 = PandasMol2().read_mol2('fragments/1_bases/in/adenosine.mol2')
RU_mol2 = PandasMol2().read_mol2('fragments/1_bases/in/uridine.mol2')
RG_mol2 = PandasMol2().read_mol2('fragments/1_bases/in/guanosine.mol2')
RC_mol2 = PandasMol2().read_mol2('fragments/1_bases/in/cytidine.mol2')
DA_rtp = pd.read_csv('forcefields/1_amber_rtp/dna.rtp', skiprows=89, sep='\s+', nrows=32, comment=';', names=['atom_name', 'atom_type', 'charge', 'nr'], na_filter=False)
DT_rtp = pd.read_csv('forcefields/1_amber_rtp/dna.rtp', skiprows=393, sep='\s+', nrows=32, comment=';', names=['atom_name', 'atom_type', 'charge', 'nr'], na_filter=False)
DG_rtp = pd.read_csv('forcefields/1_amber_rtp/dna.rtp', skiprows=700, sep='\s+', nrows=33, comment=';', names=['atom_name', 'atom_type', 'charge', 'nr'], na_filter=False)
DC_rtp = pd.read_csv('forcefields/1_amber_rtp/dna.rtp', skiprows=1007, sep='\s+', nrows=30, comment=';', names=['atom_name', 'atom_type', 'charge', 'nr'], na_filter=False)
RA_rtp = pd.read_csv('forcefields/1_amber_rtp/rna.rtp', skiprows=92, sep='\s+', nrows=33, comment=';', names=['atom_name', 'atom_type', 'charge', 'nr'], na_filter=False)
RU_rtp = pd.read_csv('forcefields/1_amber_rtp/rna.rtp', skiprows=397, sep='\s+', nrows=30, comment=';', names=['atom_name', 'atom_type', 'charge', 'nr'], na_filter=False)
RG_rtp = pd.read_csv('forcefields/1_amber_rtp/rna.rtp', skiprows=693, sep='\s+', nrows=34, comment=';', names=['atom_name', 'atom_type', 'charge', 'nr'], na_filter=False)
RC_rtp = pd.read_csv('forcefields/1_amber_rtp/rna.rtp', skiprows=1009, sep='\s+', nrows=31, comment=';', names=['atom_name', 'atom_type', 'charge', 'nr'], na_filter=False)
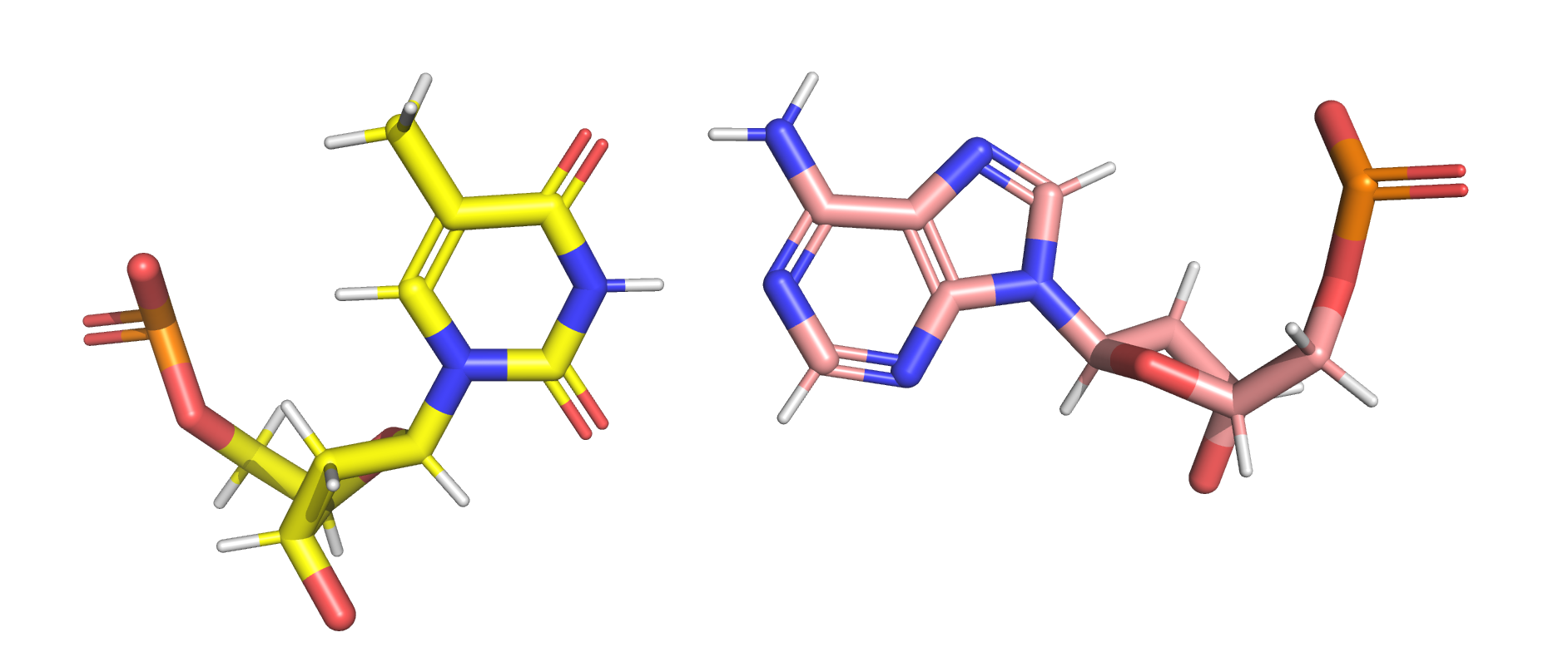
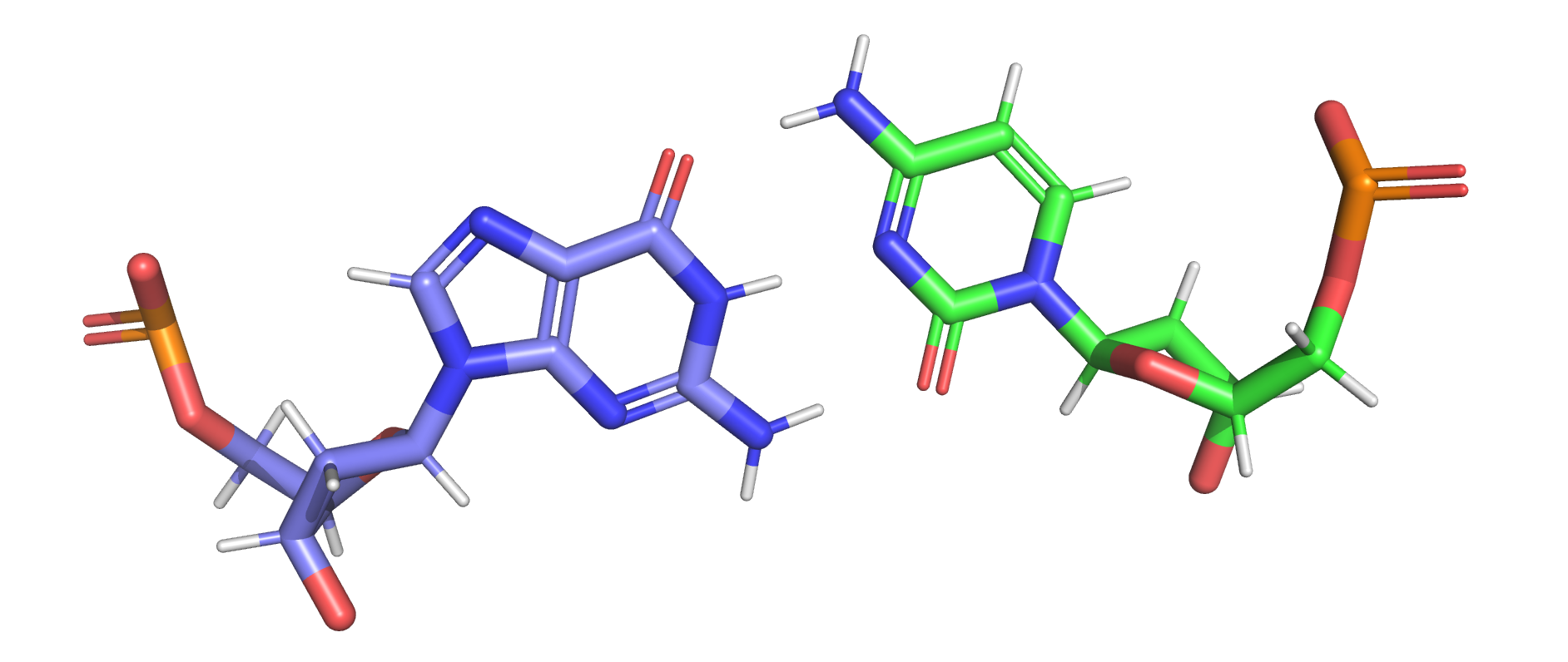
Use the fl.ff.pandasMol2_replace() function to add the charge column of the rtp file into the mol2 file. The atom_name column is used as the comparing index.
fl.ff.pandasMol2_replace(DA_rtp, DA_mol2.df, 'charge')
fl.ff.pandasMol2_replace(DT_rtp, DT_mol2.df, 'charge')
fl.ff.pandasMol2_replace(DG_rtp, DG_mol2.df, 'charge')
fl.ff.pandasMol2_replace(DC_rtp, DC_mol2.df, 'charge')
fl.ff.pandasMol2_replace(RA_rtp, RA_mol2.df, 'charge')
fl.ff.pandasMol2_replace(RU_rtp, RU_mol2.df, 'charge')
fl.ff.pandasMol2_replace(RG_rtp, RG_mol2.df, 'charge')
fl.ff.pandasMol2_replace(RC_rtp, RC_mol2.df, 'charge')
Finally, save the updated mol2 files.
fl.ff.write_mol2(DA_mol2, 'fragments/1_bases/out/deoxyadenosine.mol2')
fl.ff.write_mol2(DT_mol2, 'fragments/1_bases/out/deoxythymidine.mol2')
fl.ff.write_mol2(DG_mol2, 'fragments/1_bases/out/deoxyguanosine.mol2')
fl.ff.write_mol2(DC_mol2, 'fragments/1_bases/out/deoxycytidine.mol2')
fl.ff.write_mol2(RA_mol2, 'fragments/1_bases/out/adenosine.mol2')
fl.ff.write_mol2(RU_mol2, 'fragments/1_bases/out/uridine.mol2')
fl.ff.write_mol2(RG_mol2, 'fragments/1_bases/out/guanosine.mol2')
fl.ff.write_mol2(RC_mol2, 'fragments/1_bases/out/cytidine.mol2')