Coupling chemistries#
The design principles of FRETlabel for in silico labeling closely follow the building blocks involved when labeling a nucleic acid in vitro. FRETlabel treats the base, a carbon linker (currently only C6) and the dye moiety as individual fragments to achieve modularity. The PyMOL plugin readily implements various attachement chemistries described in the literature [3, 8, 12]. It comes with a preconfigured set of fluorophores (cyanines, Alexa and Atto dyes). Additional base-linkers fragments and fluorophores can be added by the user. A step-by-step procedure is described in the section Fragment building.
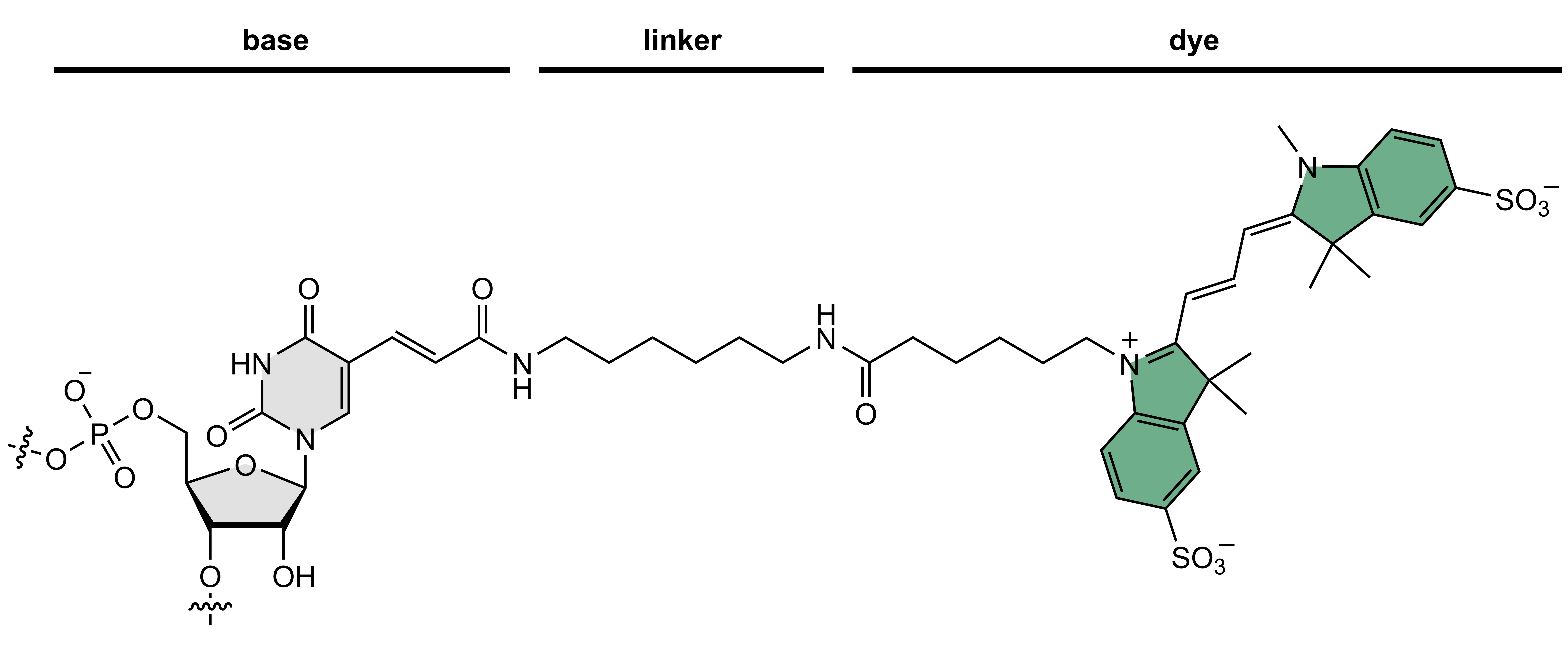
Fig. 4 A Cy3 fluorophore covalently attached to a deoxythimidine. The construct is composed of (i) a base, (ii) a linker and (iii) a dye fragment.#
End-labeling#
Nucleic acids can be labeled at the 3’- or 5’-ribose via a bridging phosphate. Additionally, insertion of a hydrazide-functionalized dye linker into the 3’-terminal sugar has been described as a way to post-transcriptionally label long RNAs [3, 12].
Internal labeling#
Internal labeling of chemically synthesized RNA and DNA fragments typically use a C5-modified deoxythimidine residue. For long RNAs ethenoadducts can be post-transcriptionally incorporated on adenosine or cytidine residues [3, 13, 14].
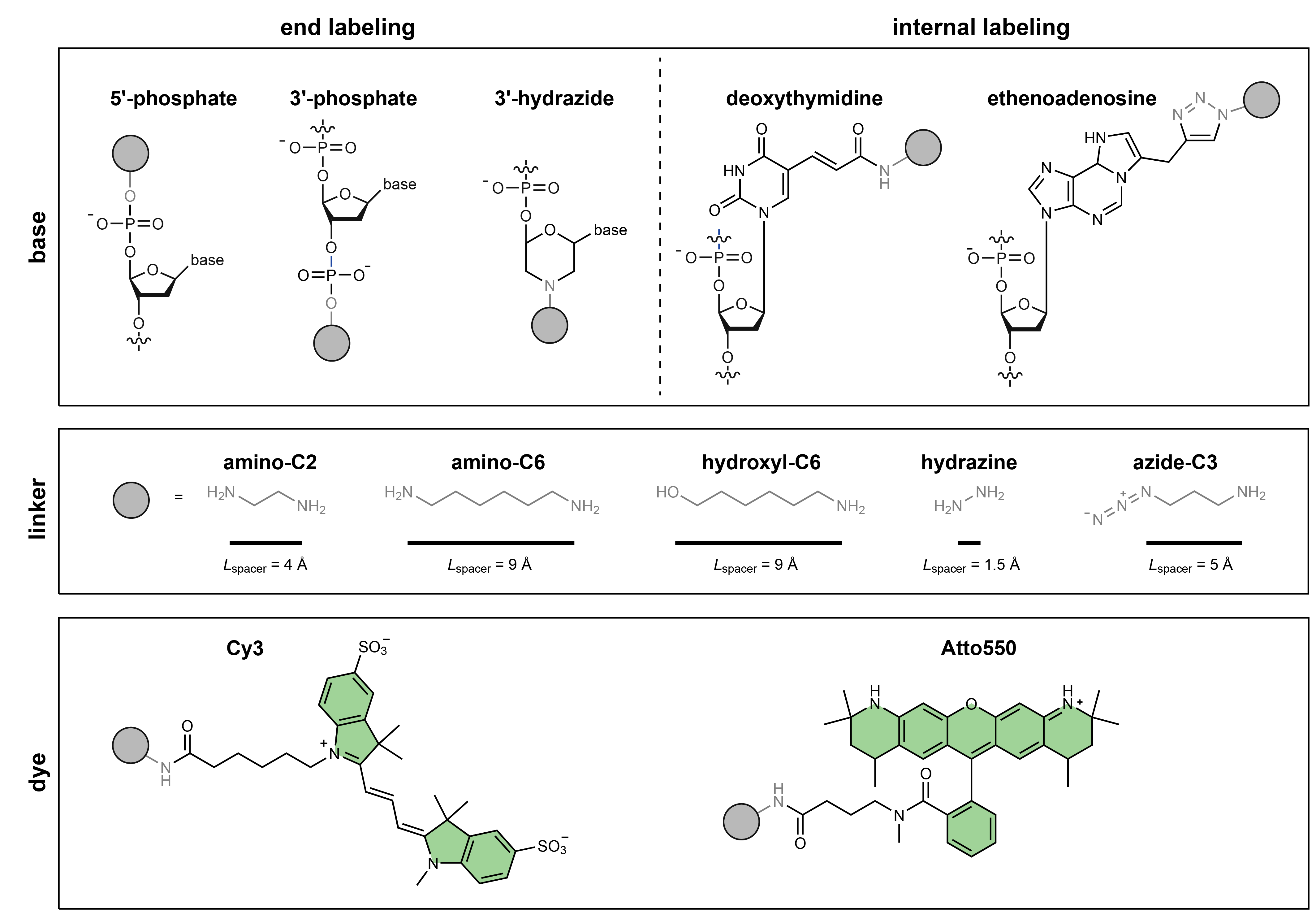
Fig. 5 Schematic of different terminal and internal coupling chemistries for labeling nucleic acids (adapted from Steffen, Bioinformatics 2021 [15]).#