Labeling with the PyMOL plugin#
FRETlabel implements the task of attaching a fluorescent dye molecule to a nucleic acid (RNA or DNA) of interest in the form of a PyMOL plugin. To get started load a PDB structure and select a target residue by chain and resi identifiers. The currently selected residue is highlighted in orange.
Tip
To help you select the target residue you may open the integrated notepad by clicking on show Text
Next, define your dye fragment by choosing the position (internal, 3’-end, 5’-end), the type of chemistry (phosphate, U/dT-C5, ethenoadduct), the base and the dye.
Finally, click on Add fluorophore to attach the dye to the biomolecule. You may save the labeled construct to a directoy of choice.
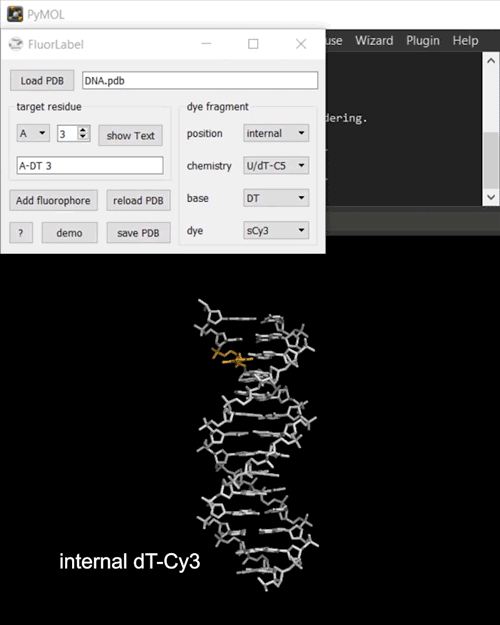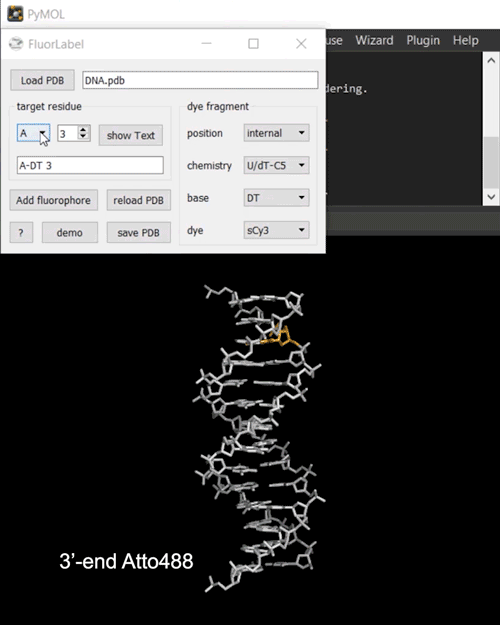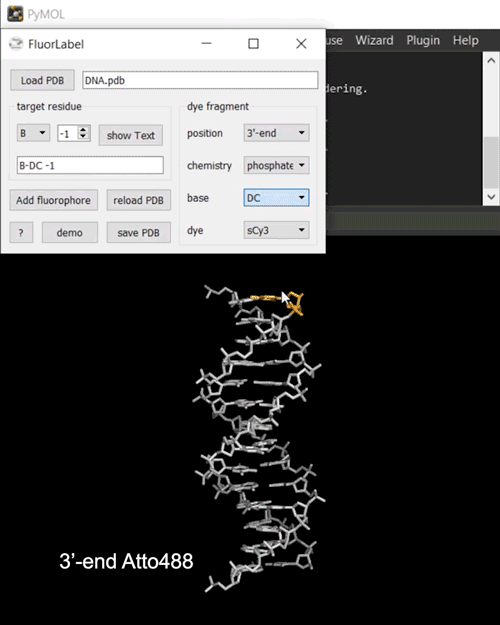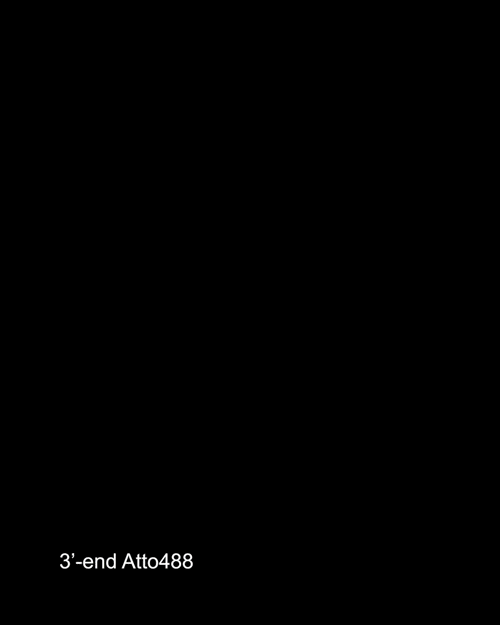
Fig. 2 Interactive fluorescence labeling of nucleic acids. (i) A Cy3 fluorophore is coupled covalently to an internal deoxythymidine. (ii) An Atto488 fluorphore is attached to the phosphorylated 3’-terminal deoxycytidine.#
Note on buried residues
In some situations the residue of choice may be inaccessible or the attached dye may clash with the biomolecule. In this case, it is necessary to manually adjust the dihedral angles of a few atoms. This can be achieved in PyMOL by going into Edit mode and ctrl + <right-clicking> on a bond. Note that relieving obvious atomic clashes in this way is a necessary first step but no replacement of a proper energy minimization (EM). Try to keep the manual modifications of the structure to a minimum and always run an EM before starting your MD simulation.