References#
Citation
If you use FRETraj in your work please cite the following paper:
F.D. Steffen, R.K.O. Sigel, R. Börner, Bioinformatics 2021, 37, 3953–3955.
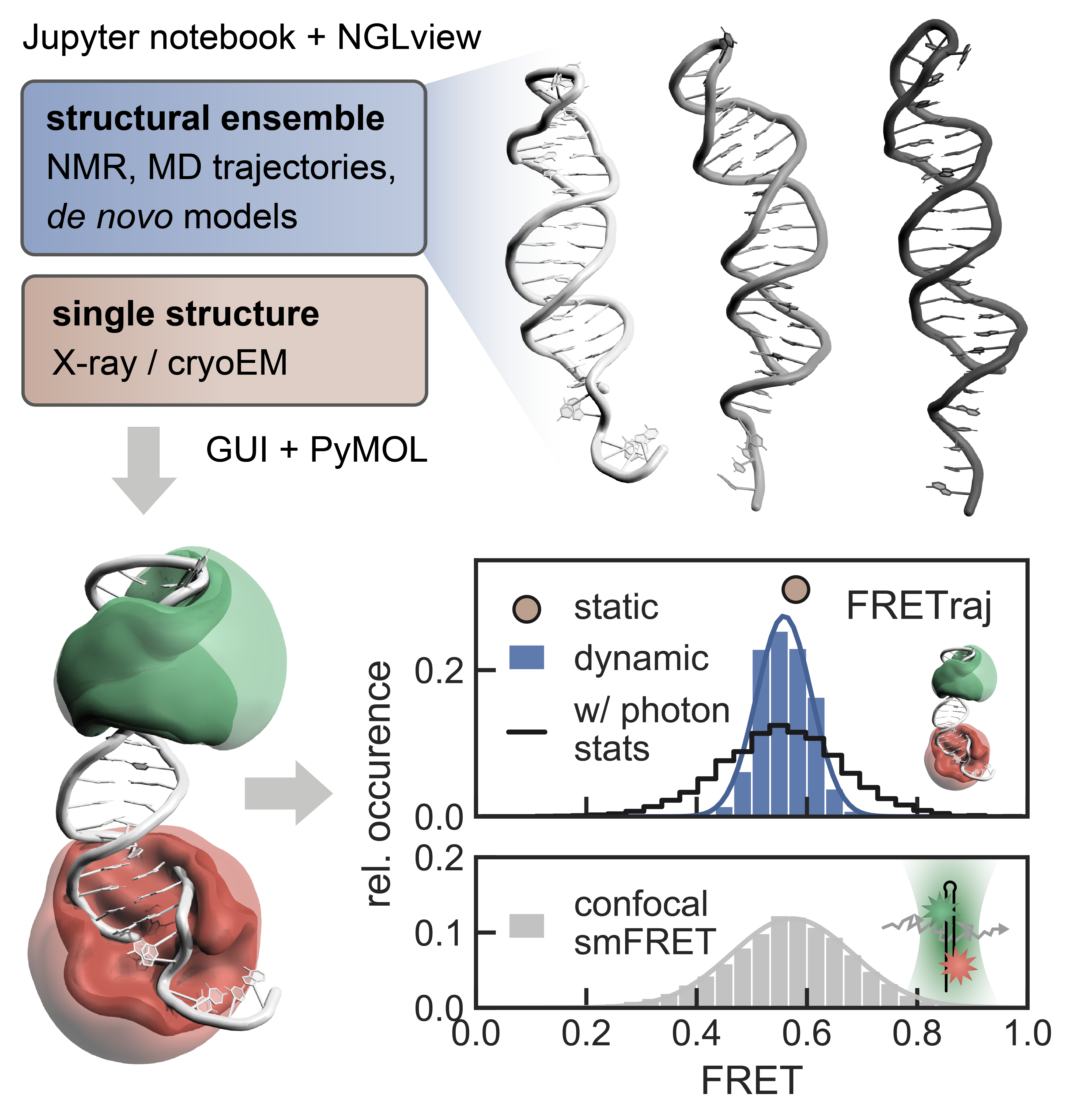
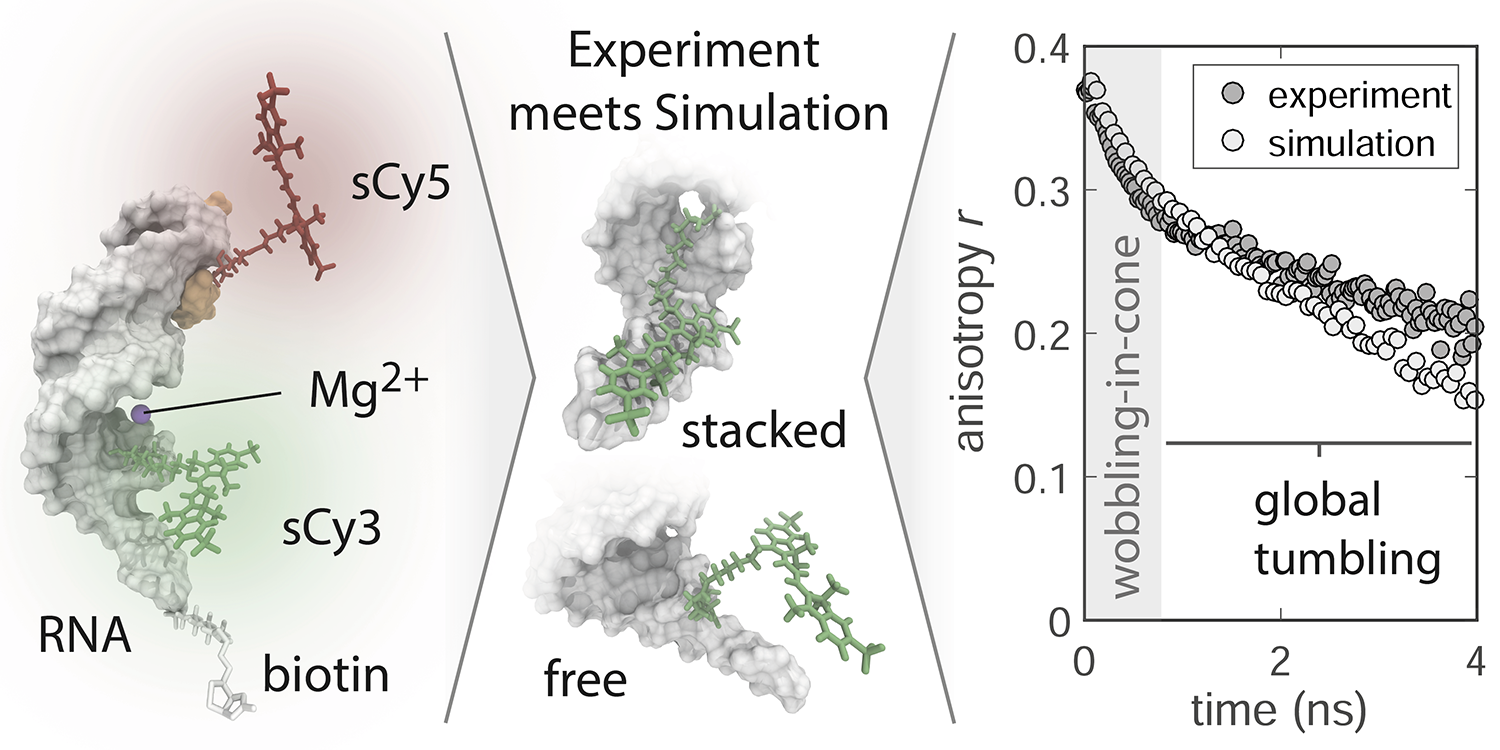
The contact volume (CV) as an extension of the accessible volume (AV) was introduced in:
F.D. Steffen, R.K.O. Sigel, R. Börner, Phys. Chem. Chem. Phys 2016, 18, 29045-29055.
Bibliography#
Warren L. DeLano. Pymol: an open-source molecular graphics tool. CCP4 Newsletter on protein crystallography, 40(1):82–92, 2002.
Thomas Kluyver, Benjamin Ragan-Kelley, Fernando Pérez, Brian Granger, Matthias Bussonnier, Jonathan Frederic, Kyle Kelley, Jessica Hamrick, Jason Grout, Sylvain Corlay, Paul Ivanov, Damián Avila, Safia Abdalla, Carol Willing, and Jupyter development team. Jupyter notebooks – a publishing format for reproducible computational workflows. In Fernando Loizides and Birgit Schmidt, editors, Positioning and Power in Academic Publishing, pages 87–90. Ios Press Inc, Amsterdam, 2016. doi:10.3233/978-1-61499-649-1-87.
Hai Nguyen, David A. Case, and Alexander S. Rose. Nglview-interactive molecular graphics for jupyter notebooks. Bioinformatics, 34(7):1241–1242, 2018. doi:10.1093/bioinformatics/btx789.
Simon Sindbert, Stanislav Kalinin, Hien Nguyen, Andrea Kienzler, Lilia Clima, Willi Bannwarth, Bettina Appel, Sabine Müller, and Claus A. M. Seidel. Accurate distance determination of nucleic acids via förster resonance energy transfer: implications of dye linker length and rigidity. J. Am. Chem. Soc., 133(8):2463–2480, 2011. doi:10.1021/ja105725e.
Björn Hellenkamp, Sonja Schmid, Olga Doroshenko, Oleg Opanasyuk, Ralf Kühnemuth, Soheila Rezaei Adariani, Benjamin Ambrose, Mikayel Aznauryan, Anders Barth, Victoria Birkedal, Mark E. Bowen, Hongtao Chen, Thorben Cordes, Tobias Eilert, Carel Fijen, Christian Gebhardt, Markus Götz, Giorgos Gouridis, Enrico Gratton, Taekjip Ha, Pengyu Hao, Christian A. Hanke, Andreas Hartmann, Jelle Hendrix, Lasse L. Hildebrandt, Verena Hirschfeld, Johannes Hohlbein, Boyang Hua, Christian G. Hübner, Eleni Kallis, Achillefs N. Kapanidis, Jae-Yeol Kim, Georg Krainer, Don C. Lamb, Nam Ki Lee, Edward A. Lemke, Brié Levesque, Marcia Levitus, James J. McCann, Nikolaus Naredi-Rainer, Daniel Nettels, Thuy Ngo, Ruoyi Qiu, Nicole C. Robb, Carlheinz Röcker, Hugo Sanabria, Michael Schlierf, Tim Schröder, Benjamin Schuler, Henning Seidel, Lisa Streit, Johann Thurn, Philip Tinnefeld, Swati Tyagi, Niels Vandenberk, Andrés Manuel Vera, Keith R. Weninger, Bettina Wünsch, Inna S. Yanez-Orozco, Jens Michaelis, Claus A. M. Seidel, Timothy D. Craggs, and Thorsten Hugel. Precision and accuracy of single-molecule fret measurements-a multi-laboratory benchmark study. Nat. Methods, 15(9):669–676, 2018. doi:10.1038/s41592-018-0085-0.
Martin Hoefling, Nicola Lima, Dominik Haenni, Claus A. M. Seidel, Benjamin Schuler, and Helmut Grubmüller. Structural heterogeneity and quantitative fret efficiency distributions of polyprolines through a hybrid atomistic simulation and monte carlo approach. PLoS One, 6(5):e19791, 2011. doi:10.1371/journal.pone.0019791.
Martin Hoefling and Helmut Grubmüller. In silico fret from simulated dye dynamics. Comput. Phys. Commun., 184(3):841–852, 2013. doi:10.1016/j.cpc.2012.10.018.
Fabio D. Steffen, Roland K. O. Sigel, and Richard Börner. An atomistic view on carbocyanine photophysics in the realm of rna. Phys. Chem. Chem. Phys., 18(42):29045–29055, 2016. doi:10.1039/c6cp04277e.
Stanislav Kalinin, Thomas Peulen, Simon Sindbert, Paul J. Rothwell, Sylvia Berger, Tobias Restle, Roger S. Goody, Holger Gohlke, and Claus A. M. Seidel. A toolkit and benchmark study for fret-restrained high-precision structural modeling. Nat. Methods, 9(12):1218–1225, 2012. doi:10.1038/nmeth.2222.










